May, 2024
"Background: To identify genotypes associated with neovascular age-related macular degeneration (nAMD) and investigate the associations between genotype variations and anti-vascular endothelial growth factor (VEGF) treatment response.
Methods: This observational, retrospective, case series study enrolled patients diagnosed with nAMD who received anti-VEGF treatment in National Taiwan University Hospital with at least one-year follow-up between 2012 and 2020. A genome-wide association study (GWAS) was conducted on enrolled patients and controls. Correlations between the genotypes identified from GWAS and the treatment response of functional/anatomical biomarkers, including visual acuity (VA), presence of intraretinal or subretinal fluid (SRF), serous or fibrovascular pigmented epithelium detachment (PED), and disruption of the ellipsoid zone (EZ), were analysed.
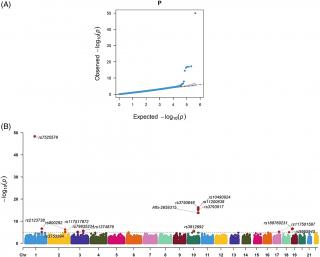
Results: In total, 182 patients with nAMD and 1748 controls were enrolled. GWAS revealed 16 single nucleotide polymorphisms (SNPs) as risk loci for nAMD, including seven loci in CFH and ARMS2/HTRA1 and nine novel loci, including rs117517872 and rs79835234(COPB2-DT), rs7525578(RAP1A), rs2123738(LOC105376755), rs1374879(CNTN3), rs3812692(SAR1A), rs117501587(PRKCA), rs9965945(CNDP1), and rs189769231(MATK). Our study revealed rs800292(CFH), rs11200638(HTRA1), and rs2123738(LOC105376755) correlated with poor treatment response in VA (P = 0.005), SRF (P = 0.044), and fibrovascular PED (P = 0.007), respectively. Rs9965945(CNDP1) was correlated with poor response in disruption of EZ (P = 0.046) and serous PED (P = 0.049).
Conclusions: Among the 16 SNPs found in the GWAS, four loci-CFH, ARMS2/HTRA1, and two novel loci-were correlated with the susceptibility of nAMD and anatomical/functional responses after anti-VEGF treatment."